Recently, a review article entitled “Expanding the plant epigenetic code: histone short-chain acylation” was published in the international journal Trends in Plant Science by the research team led by Xu Qiutao and Zhang Jisen from the School of Agriculture. The article systematically elaborates on the functional mechanisms of histone short-chain acylation. Wei Xuelu, a PhD candidate at the School of Agriculture, and Shao Guiyu, a master's student, are co–first authors. Professor Chen Xiaoyang of Anhui Agricultural University, and Professors Xu Qiutao and Zhang Jisen of the GXU School of Agriculture serve as co–corresponding authors. GXU is the first-completing institution of the article.
Histone methylation and acetylation are classical epigenetic marks that play central roles in regulating chromatin structure and gene expression. In recent years, with the rapid development of high-resolution mass spectrometry, multiple novel short-chain acylation modifications on histone lysine residues have been continuously identified, including crotonylation (Kcr), butyrylation (Kbu), β-hydroxybutyrylation (Kbhb), 2-hydroxyisobutyrylation (Khib), succinylation (Ksu), and lactylation (Kla). The discovery of these new types of modifications greatly enriches the repertoire of histone post-translational modifications and provides new perspectives for understanding epigenetic regulatory mechanisms. These short-chain acylation marks are primarily catalyzed by histone acyltransferases (“writers”) and dynamically regulated by histone deacylases (“erasers”). Studies in animals suggest that these acyl–CoA donors typically originate from specific metabolic pathways. In plants, although the precise metabolic sources of many acyl donors remain unclear, current evidence indicates that they may be derived from fatty acid β-oxidation, the tricarboxylic acid (TCA) cycle, and glycolysis (Figure 1).
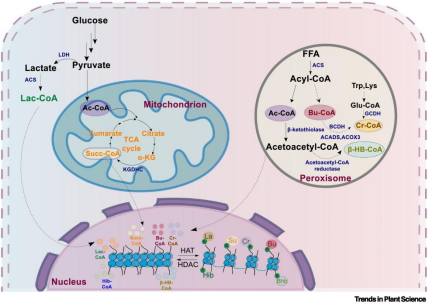
Figure 1. Metabolic pathways of acyl–CoA in plant cells
As a highly dynamic class of epigenetic modifications, lysine short-chain acylation has been shown to play widespread regulatory roles in plant growth, development, and stress adaptation. Research has demonstrated that these modifications are broadly distributed across various plant tissues. For example, Khib is widespread in wheat leaves and rice flowers; Ksu is abundant in leaves and roots; and Kla has been detected in wheat and rice seeds. These modifications influence plant growth and development by regulating key biological processes such as central carbon metabolism, photosynthesis, protein synthesis, and nutrient storage. In terms of stress adaptation, lysine short-chain acylation also plays critical regulatory roles. Studies show that such modifications can respond to multiple environmental stresses, including starvation, drought, salinity, low temperature, and pathogen infection. For instance, histone Kcr and Kbu dynamically participate in rice responses to starvation and flooding stress, while pathogen infection can induce the H4K8hib modification in rice, thereby promoting the expression of immunity-related genes (Figure 2).
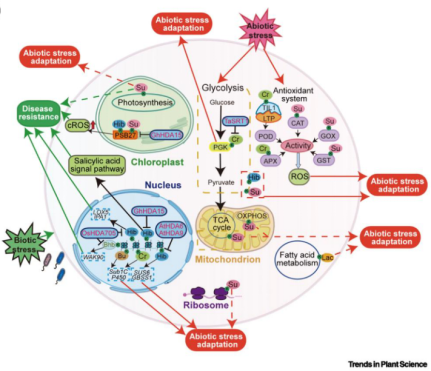
Figure 2. Molecular mechanisms by which lysine short-chain acylation regulates plant development and stress
adaptation
The review also provides future perspectives for research in this field, highlighting key scientific questions such as the interplay among different acylation modifications, the metabolic regulatory networks underlying plant histone acylation, and the identification of acylation “reader” proteins. Breakthroughs in these research directions will not only enrich and advance epigenetic theory but also open new avenues and innovative strategies for breeding stress-resistant crops.
This work was supported by the National Key R&D Program, major projects fund of Guangxi, the National Natural Science Foundation of China, the Guangxi Distinguished Young Scholars Fund, and the High-level Talent Program of Guangxi University.